Abstract
Next-Generation Sequencing (NGS) has recently been introduced to efficiently and simultaneously detect genetic variations in acute myeloid leukemia (AML). However, its implementation in the clinical routine raises new challenges focused on the diversity of assays and variant reporting criteria. To overcome this challenge, the PETHEMA group established a nationwide network of 7 reference laboratories aimed to deliver molecular results to the clinics. We report the technical cross-validation results for NGS and clinical validation in 2960 AML samples.
Three cross-validation rounds (CVR) were performed to establish consensus parameters for NGS analysis and variant reporting. In the first CVR we evaluated the starting situation of the NGS studies. In the second CVR the laboratory network established minimum quality parameters and consensus recommendations to guarantee a valid NGS assay. The third CVR strengthened the established parameters and refined the clinical variant classification. The clinical validation was performed in 2960 samples from 2703 patients: 1530 male and 1173 female with a median age of 67.5 years old. 2522 samples were collected at diagnosis, 275 at relapse and 163 at refractory AML from October 2017 to October 2019. NGS analysis was performed according to already implemented protocols and only variants accomplishing the established quality control parameters were considered.
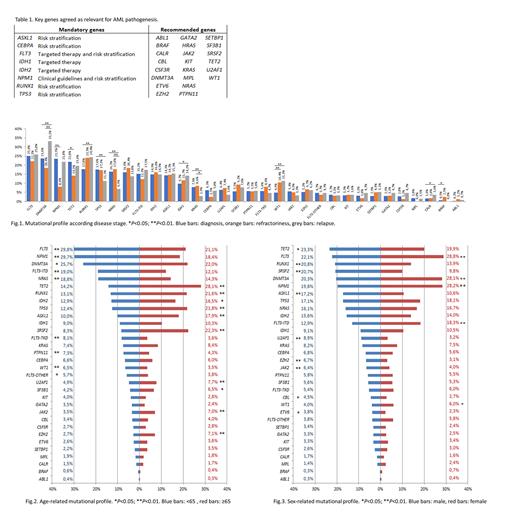
In the first CVR the error rate (ER) was 39% with a high variability in the studied genes. Then, 30 genes were agreed as key genes for AML pathogenesis: 8 were considered mandatory due to their implication in clinical guidelines, targeted therapy and risk stratification and the study of the remaining 22 was recommended based on panel availability (Table 1). In the second CVR the ER was reduced to 14.4% and the NGS quality metrics were 4032X of mean read depth and 98.3% of median uniformity. Therefore, the laboratory network established a minimum read depth of 500X and uniformity >85% as quality control parameters. Due to the high variability in the detection of low VAF variants (1 CVR: ER<5%: 86.1%; 2 CVR: ER<5%: 28.6%), VAF≥5% was established as a cut-off for variant reporting excepting variants with strong clinical evidence. In the third CVR the error rate for VAF≥5% variants was 10.9% achieving high concordance in variant detection and clinical classification.
8043 variants were reported in the 2960 samples, with 96.5% of patients showing at least 1 mutated gene. The mean number of variants per sample was 2.71 (range 0-9), the number of mutations in ≥65 years old patients (2.9 vs. 2.5, P<0.001) and in male patients (2.8 vs. 2.6, P=0.001) were significantly higher. In the global cohort, the most frequently mutated genes were FLT3 (24.9%), DNMT3A (23.9%) and NPM1 (23%). However, this genetic profile changed according to the disease stage, age and sex. NPM1 mutations were more frequent at diagnosis (P<0.001), IDH1 mutations (P=0.027) RUNX1 mutations (P=0.011) and WT1 mutations (P<0.001) were more frequent at relapse and mutations in KRAS (P=0.012) were more frequently detected in refractory AML (Fig 1). Younger patients (<65) had more mutations in FLT3 and NPM1 (P<0.001) while mutations in ASXL1, EZH2, IDH2, JAK2, RUNX1, SRSF2, TET2, TP53, U2AF1 and SF3B1 were associated to older age (P<0.05) (Fig 2). NPM1, FLT3 and DNMT3A were frequently mutated in female patients (P<0.001) while male patients had more mutations in ASXL1, JAK2, EZH2, RUNX1, SRSF2 and U2AF1 (P<0.01) (Fig 3).
At least 1 mutation in one of the 8 clinically relevant genes was detected in the 78.7% of patients: 39.8% of patients had targetable mutations (FLT3-ITD/TKD and IDH1/2 mutations) and 35.6% of patients, variants found in ASXL1, RUNX1 and TP53 were the only clinically relevant variant detected. The study of clonal evolution through paired-sample analysis showed that mutations in FLT3, KRAS, NRAS and PTPN11 were particularly unstable at relapse or refractoriness.
We show the development of the first national strategy for validation of NGS studies with centralized analysis in an AML cooperative group. We have developed a laboratory network with standardized protocols to ensure technical quality and equity in access to NGS studies. The unification of analysis and interpretation criteria represents a significant increase in the quality of diagnostic tests and translational research.
N/A-NI-AML-PETHEMA-007343, PI18/01340, PI19/00730, FI19/00059
Ayala: Incyte Corporation: Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees; Astellas: Honoraria; Celgene: Honoraria. Tormo: Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Jazz Pharmaceuticals: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Pfizer: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Amgen: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Astellas: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau. Perez-Simon: JANSSEN, TAKEDA, PFIZER, JAZZ, BMS, AMGEN, GILEAD: Other: honorarium or budget for research projects and/or participation in advisory boards and / or learning activities and / or conferences. Martínez-López: Janssen, BMS, Novartis, Incyte, Roche, GSK, Pfizer: Consultancy; Roche, Novartis, Incyte, Astellas, BMS: Research Funding. Montesinos: AbbVie: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Astellas: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Agios: Consultancy; Celgene: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Daiichi Sankyo: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Incyte: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Janssen: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Karyopharm: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Pfizer: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Sanofi: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Teva: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Forma Therapeutics: Consultancy; Glycomimetics: Consultancy; Tolero Pharmaceutical: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal